The building plan
Storing the building plans for a virus in its genome is much like how we store ideas in language. This may sound strange but, as an example, typos in spelling, grammar, or word usage, can lead to the meaning of a sentence either changing dramatically, remaining virtually unchanged, or becoming complete nonsense. The SARS-CoV-2 genome consists of RNA. Transcription of this RNA runs into a similar problem: errors can lead to the loss of function, a gain of function, or be completely inconsequential to the resulting protein (Figure 1). Large changes may break the virus, but smaller changes may provide an advantage and are essential for evolution.
Targeting the copy machine
In a previous article we spoke about the copy machinery of the virus, including the RNA-dependent RNA polymerase (RdRp), and drugs targeting it, such as Remdesivir. The goal of these drugs is to jam the enzyme and halt RNA production - or to cause more errors than are sustainable, with the end result being a less infectious virus. The reason the development of drugs targeting the copy machinery of RNA is worthwhile is that humans don’t have machinery to reproduce RNA from RNA. This means drugs targeting this machinery are less likely to interfere with normal processes in people. What if the virus could quickly repair these errors before the new genome is packed into a hull and kicked out the door? That would make finding a therapeutic much more difficult…
Correctional facilities
Unfortunately, SARS-CoV-2 has a way to repair the mistakes. When errors are introduced in transcription through environmental mutagenesis or even mutations caused by nucleotide analogs like Ribavarin1–3, the non-structural protein 14 (nsp14) has the ability to remove them. This multifunctional protein removes errors with the exoribonuclease (ExoN) activity of its N-terminal domain, while the C-terminal domain has the unrelated function of methylating the end cap of the viral RNA3,4.
However, this ExoN does not work alone. There is a replication complex made up of proteins performing many roles in the production of new RNA with high fidelity. Nsp12 is the main hub that makes a new RNA chain to complement the template. Nsp7 and nsp8 have a “processivity” role to enable nsp12 to function efficiently. In addition to these proteins there is a two-component proofreading system of Helicase (nsp13) and the ExoN domain of nsp14. Helicase can detect misshapen RNA helices caused by errors made by the copy machinery5. It then unwinds these double strands of RNA and feeds the strand containing the error into the ExoN domain of nsp14 where they are chopped out. This results in nsp12 continuing RNA replication where it left off.
Exoribonuclease or no exoribonuclease
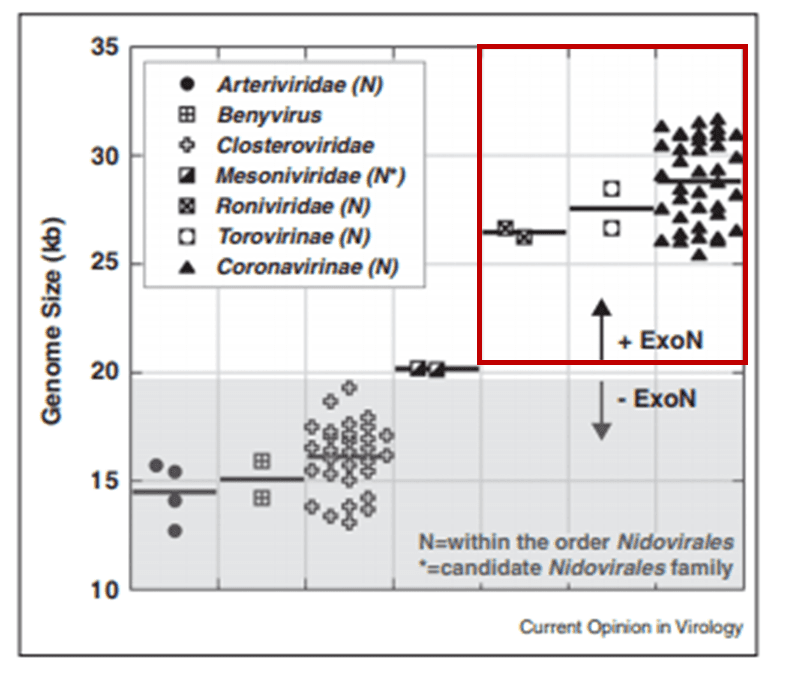
The proofreading ability from Helicase and nsp14 ExoN allows SARS-CoV-2 to have a huge genome as compared to other viruses6(Figure 2). The large 29.9 kb genome of SARS-CoV-2 requires much more physical space to accommodate the necessary genetic information for reproduction when compared to other RNA viruses, such as Rhinovirus that has a genome between 7.2 kb and 8.5 kb in size (Figure 3). When no ExoN proofreading is present genomes cannot expand beyond 20 kb in size6(Figure 2). Maybe by removing the exoribonuclease activity, irreversible damage could be caused to the genome of SARS-CoV-2.

Nsp14 Structure
In order to understand how nsp14 can do this, we need to find out its atomic structure; this may also allow us to develop a drug which hinders its function. However, to this date, no structure of nsp14 from SARS-CoV-2 has been solved. However, structures have been solved of nsp14 in complex with another viral protein, nsp10, both from SARS-CoV (PDB entries 5nfy, 5c8s, 5c8t, 5c8u)2,7. As the protein sequences are very similar between SARS-CoV and SARS-CoV-2 (nsp14 is 95%, and nsp10 is 97% identical), it can be assumed that the SARS-CoV-2 structure as well as its functionality are very similar to SARS-CoV. The active site of the ExoN domain of nsp14 from SARS-CoV-2 has a DEEDh motif (named for the one-letter codes of the amino acids involved) containing a histidine as well as two aspartates and two glutamates2,3,7,8.
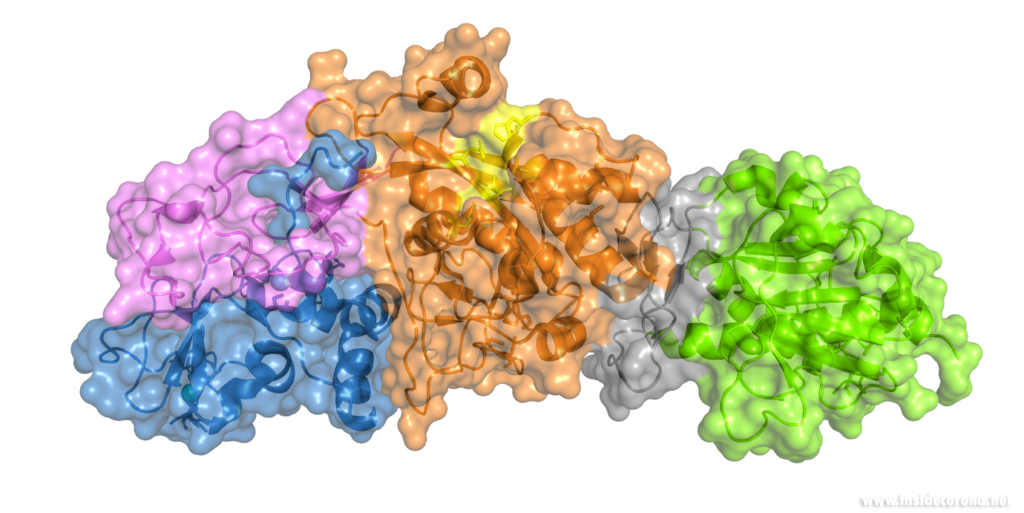
Nsp14 interacts with nsp10
The N-terminus of nsp14 interacts with nsp10 (pink and blue, respectively, in Figure 4). The following domain (orange) has been shown to have exoribonuclease activity on double stranded RNA in a 3’ to 5’ direction9. When nsp10 is interacting with nsp14 there is a 35 fold increase in exoribonuclease activity, which is thought to occur due to conformational changes caused by formation of the complex2,9. The ExoN domain of nsp14 (orange) is connected to the methyltransferase domain (green) by a flexible hinge (black)7,10. This flexible region opens up the methyltransferase active site to allow methylation of the N7 of the 5’ Guanosine triphosphate of RNA10. There are three zinc finger motifs in nsp14 with two found in the ExoN domain and one in the methyltransferase domain2,7. In combination with the two further zinc sites in nsp10, these zinc fingers hold loops of the proteins together and are involved with nucleotide interaction2,7.
Nsp14 has also been demonstrated to form complexes with the copy machinery , nsp12, nsp7, and nsp8, although this interaction is independent of nsp102,11,12.
Exoribonuclease active site and potential drug development
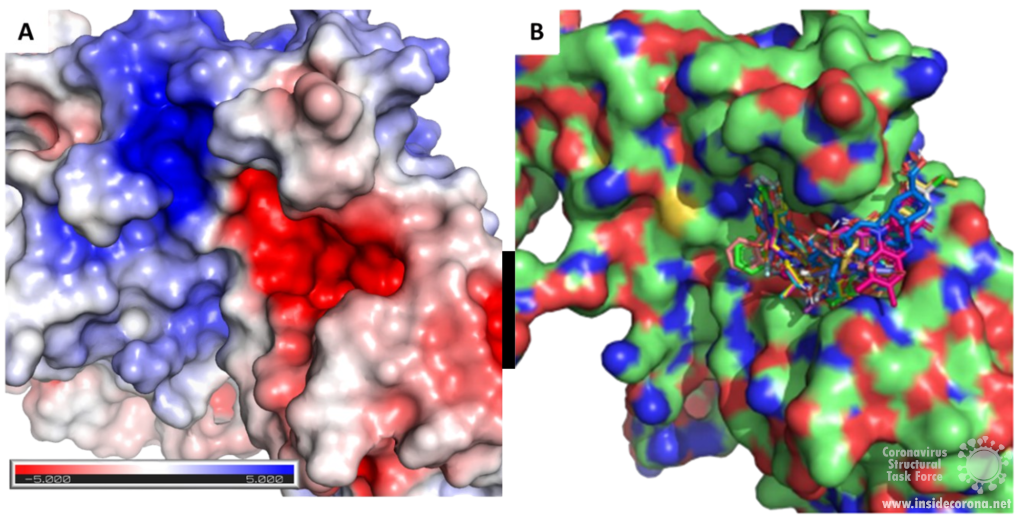
Scientists are searching for drugs that could be used to target nsp14 in order to find a cure for COVID-19. The active site of the ExoN domain of nsp14 has five residues that are essential for activity that form a negatively charged pocket (Figure 5A)7. Currently researchers are using the nsp14 structure from SARS-CoV to model a SARS-CoV-2 structure which can be used to identify compounds that could bind to the active site (Figure 5). These in silico screens start with nucleotide analog drugs like Remdesivir, Ribivarin or Ritonavir that are currently used as antiviral treatments for other viruses13–15. These nucleotide analogs are then changed to achieve a better binding to Nsp14’s active site in order to block it (Figure 5B).
As the ExoN is essential to support the huge 29.9kb genome of SARS-CoV-2, targeting nsp14 could lead to an effective treatment to COVID-19. Although drugs that target just nsp14 could be effective at increasing the error rate in RNA production by the virus, a more effective treatment will require inhibition of the RdRp of the copy machinery at the same time!
Available structures
If you would like to look at the currently available structures for Nsp14(currently only available from SARS-CoV), they are available from our data base; we provide information on the quality of measurement data and models as well as improved structures. The highest resolution structure of nsp14 is PDB entry 5c8t at 3.2Å. This has a bound S-Adenosyl methionine ligand as well as zinc atoms present. Alongside this, another structure of Nsp14 bound to S-Adenosyl homocysteine and a guanosine-triphosphate-adenosine ligand as well as zinc at 3.33Å resolution has been published (PDB: 5c8s). Additionally, two structures with zinc atoms but no ligands are available (PDB 5c8u 3.4Å at and 5nfy at 3.34Å). Both PDB entry 5c8t and 5nfy have improved structures re-refined by our group.
Sources
- 1.Zuo Y. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Research. Published online March 1, 2001:1017-1026. doi:10.1093/nar/29.5.1017
- 2.Ferron F, Subissi L, Silveira De Morais AT, et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc Natl Acad Sci USA. Published online December 26, 2017:E162-E171. doi:10.1073/pnas.1718806115
- 3.Barnes MH, Spacciapoli P, Li DH, Brown NC. The 3′–5′ exonuclease site of DNA polymerase III from Gram-positive bacteria: definition of a novel motif structure. Gene. Published online January 1995:45-50. doi:10.1016/0378-1119(95)00530-j
- 4.Chen Y, Cai H, Pan J, et al. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proceedings of the National Academy of Sciences. Published online February 10, 2009:3484-3489. doi:10.1073/pnas.0808790106
- 5.Chen J, Malone B, Llewellyn E, et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Published online July 8, 2020. doi:10.1101/2020.07.08.194084
- 6.Smith EC, Denison MR. Implications of altered replication fidelity on the evolution and pathogenesis of coronaviruses. Current Opinion in Virology. Published online October 2012:519-524. doi:10.1016/j.coviro.2012.07.005
- 7.Ma Y, Wu L, Shaw N, et al. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc Natl Acad Sci USA. Published online July 9, 2015:9436-9441. doi:10.1073/pnas.1508686112
- 8.Eckerle LD, Becker MM, Halpin RA, et al. Infidelity of SARS-CoV Nsp14-Exonuclease Mutant Virus Replication Is Revealed by Complete Genome Sequencing. Emerman M, ed. PLoS Pathog. Published online May 6, 2010:e1000896. doi:10.1371/journal.ppat.1000896
- 9.Bouvet M, Imbert I, Subissi L, Gluais L, Canard B, Decroly E. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proceedings of the National Academy of Sciences. Published online May 25, 2012:9372-9377. doi:10.1073/pnas.1201130109
- 10.Ogando NS, Ferron F, Decroly E, Canard B, Posthuma CC, Snijder EJ. The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity. Front Microbiol. Published online August 7, 2019. doi:10.3389/fmicb.2019.01813
- 11.Subissi L, Posthuma CC, Collet A, et al. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc Natl Acad Sci USA. Published online September 2, 2014:E3900-E3909. doi:10.1073/pnas.1323705111
- 12.Subissi L, Imbert I, Ferron F, et al. SARS-CoV ORF1b-encoded nonstructural proteins 12–16: Replicative enzymes as antiviral targets. Antiviral Research. Published online January 2014:122-130. doi:10.1016/j.antiviral.2013.11.006
- 13.Khater S, Dasgupta N, Das G. Combining SARS-CoV-2 proofreading exonuclease and RNA-dependent RNA polymerase inhibitors as a strategy to combat COVID-19: a high-throughput in silico screen. Published online June 24, 2020. doi:10.31219/osf.io/7x5ek
- 14.Shannon A, Le NT-T, Selisko B, et al. Remdesivir and SARS-CoV-2: Structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antiviral Research. Published online June 2020:104793. doi:10.1016/j.antiviral.2020.104793
- 15.Narayanan N, Nair DT. Ritonavir May Inhibit Exoribonuclease Activity of Nsp14 from the SARS-CoV-2 Virus and Potentiate the Activity of Chain Terminating Drugs. chemrxiv.org. Published May 13, 2020. https://chemrxiv.org/articles/Ritonavir_May_Inhibit_Exoribonuclease_Activity_of_Nsp14_from_the_SARS-CoV-2_Virus_and_Potentiate_the_Activity_of_Chain_Terminating_Drugs/12280043
